🎉 New open-access article in Genome Biology!
🧪🎊 LLPS benchmarking bonanza!
We’re thrilled to announce our latest study, Comprehensive protein datasets and benchmarking for liquid–liquid phase separation studies, is now online in Genome Biology! 📘🔥
- Published: 8 July 2025
- Lead author: Carlos (huge kudos to Salva’s team!)
- 🔗 You can read it here
🌟 What’s inside?
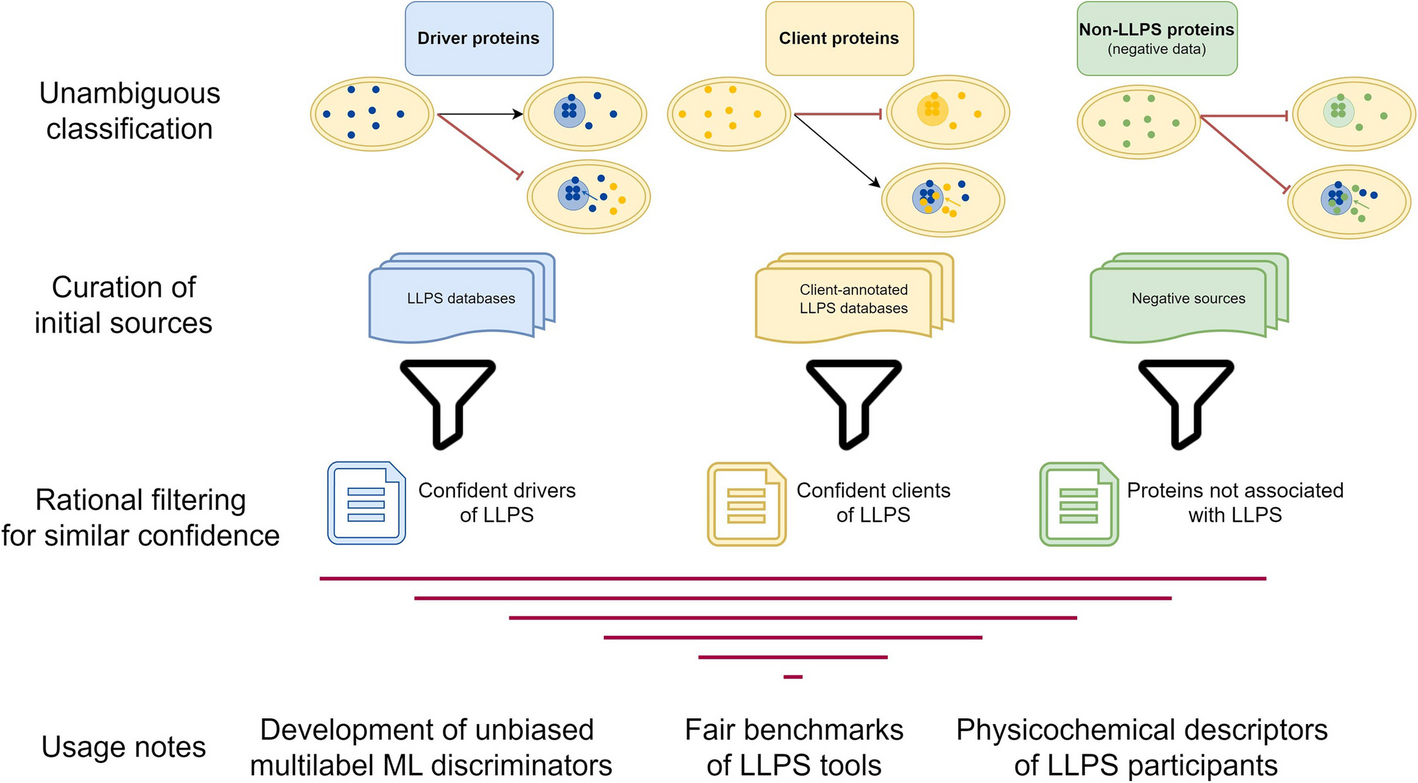
LLPS (liquid–liquid phase separation) drives biomolecular condensates, crucial for cellular function. But variability across LLPS databases makes modeling tricky. This study tackles that challenge by:
- Collecting comprehensive protein datasets across multiple LLPS repositories
- Creating benchmark frameworks to evaluate predictor performance
- Defining a reliable set of negative (non-LLPS) proteins for fair model assessment
The goal? To enable transparent, reproducible, and robust ML-driven insights into protein phase behavior.
🎯 Why it matters? 🤔
- Harmonization: brings together divergent LLPS resources for interoperability
- Model reliability: improves confidence in ML predictions with curated benchmarks
- Research enabler: helps the broader community build and validate new tools for condensate biology
By offering standardized datasets and robust benchmarking, this publication sets a new bar for reproducible ML in LLPS research.
🎉 Kudos to the team
Special shout-out to Carlos for leading the project with Oriol, Eva, Valen, Michał and Salva! Your teamwork made this possible 👏
🚀 What’s Next?
We’re already leveraging these benchmarking datasets in our ongoing LLPS and amyloid research.. If you’re into biomolecular condensates or predictive modeling, this resource is for you!
🎉 Let’s continue driving reproducible and data-driven science in LLPS research. Congrats, team! 🥳